biotite.sequence.io.genbank.GenBankFile¶
- class biotite.sequence.io.genbank.GenBankFile[source]¶
Bases:
TextFileThis class represents a file in GenBank format (including GenPept).
A GenBank file annotates a reference sequence with features such as positions of genes, promoters, etc. Additionally, it provides metadata further describing the file.
A file is divided into separate fields, e.g. the DEFINITION field contains a description of the file. The field name starts at the beginning of a line, followed by the content. A field may contain subfields, whose name is indented. For example, the SOURCE field contains the ORGANISM subfield. Some fields may occur multiple times, e.g. the REFERENCE field. A sample GenBank file can be viewed at https://www.ncbi.nlm.nih.gov/Sitemap/samplerecord.html.
This class provides a low-level interface for parsing, editing and writing GenBank files. It works like a list of field entries, where a field consists of the field name, the field content and the subfields. The field content is separated into the lines belonging to the content. While the content of metadata fields starts at the standard GenBank indentation of 12, the content of the FEATURES (contains the annotation) and ORIGIN (contains the sequence) fields starts without indentation. The subfields are represented by a dictionary, with subfield names being keys and the corresponding lines being values. The FEATURES and ORIGIN fields have no subfields.
Every entry can be obtained, set and deleted via the index operator.
Notes
This class does not support location identifiers with references to other Entrez database entries, e.g.
join(1..100,J00194.1:100..202).Examples
Create a GenBank file from scratch:
>>> file = GenBankFile() >>> file.append( ... "SOMEFIELD", ["One line", "A second line"], ... subfields={"SUBFIELD1": ["Single Line"], "SUBFIELD2": ["Two", "lines"]} ... ) >>> print(file) SOMEFIELD One line A second line SUBFIELD1 Single Line SUBFIELD2 Two lines // >>> name, content, subfields = file[0] >>> print(name) SOMEFIELD >>> print(content) ['One line', 'A second line'] >>> print(subfields) OrderedDict([('SUBFIELD1', ['Single Line']), ('SUBFIELD2', ['Two', 'lines'])])
Adding an additional field:
>>> file.insert(0, "OTHERFIELD", ["Another line"]) >>> print(len(file)) 2 >>> print(file) OTHERFIELD Another line SOMEFIELD One line A second line SUBFIELD1 Single Line SUBFIELD2 Two lines //
Overwriting and deleting an existing field:
>>> file[1] = "NEWFIELD", ["Yet another line"] >>> print(file) OTHERFIELD Another line NEWFIELD Yet another line // >>> file[1] = "NEWFIELD", ["Yet another line"], {"NEWSUB": ["Subfield line"]} >>> print(file) OTHERFIELD Another line NEWFIELD Yet another line NEWSUB Subfield line // >>> del file[1] >>> print(file) OTHERFIELD Another line //
Parsing fields from a real GenBank file:
>>> import os.path >>> file = GenBankFile.read(os.path.join(path_to_sequences, "gg_avidin.gb")) >>> print(file) LOCUS AJ311647 1224 bp DNA linear VRT 14-NOV-2006 DEFINITION Gallus gallus AVD gene for avidin, exons 1-4. ACCESSION AJ311647 VERSION AJ311647.1 GI:13397825 KEYWORDS AVD gene; avidin. SOURCE Gallus gallus (chicken) ORGANISM Gallus gallus Eukaryota; Metazoa; Chordata; Craniata; Vertebrata; Euteleostomi; Archelosauria; Archosauria; Dinosauria; Saurischia; Theropoda; Coelurosauria; Aves; Neognathae; Galloanserae; Galliformes; Phasianidae; Phasianinae; Gallus. REFERENCE 1 AUTHORS Wallen,M.J., Laukkanen,M.O. and Kulomaa,M.S. TITLE Cloning and sequencing of the chicken egg-white avidin-encoding gene and its relationship with the avidin-related genes Avr1-Avr5 JOURNAL Gene 161 (2), 205-209 (1995) PUBMED 7665080 REFERENCE 2 AUTHORS Ahlroth,M.K., Kola,E.H., Ewald,D., Masabanda,J., Sazanov,A., Fries,R. and Kulomaa,M.S. TITLE Characterization and chromosomal localization of the chicken avidin gene family JOURNAL Anim. Genet. 31 (6), 367-375 (2000) PUBMED 11167523 REFERENCE 3 (bases 1 to 1224) AUTHORS Ahlroth,M.K. TITLE Direct Submission JOURNAL Submitted (09-MAR-2001) Ahlroth M.K., Department of Biological and Environmental Science, University of Jyvaskyla, PO Box 35, FIN-40351 Jyvaskyla, FINLAND FEATURES Location/Qualifiers source 1..1224 /organism="Gallus gallus" /mol_type="genomic DNA" ... >>> name, content, _ = file[3] >>> print(name) VERSION >>> print(content) ['AJ311647.1 GI:13397825'] >>> name, content, subfields = file[5] >>> print(name) SOURCE >>> print(content) ['Gallus gallus (chicken)'] >>> print(dict(subfields)) {'ORGANISM': ['Gallus gallus', 'Eukaryota; Metazoa; Chordata; ...', ...]}
- append(name, content, subfields=None)¶
Create a new GenBank field at the end of the file.
- Parameters
- namestr
The field name.
- contentlist of str
The content lines.
- subfield_dictdict of str -> str, optional
The subfields of the field. The dictionary maps subfield names to the content lines of the respective subfield.
- copy()¶
Create a deep copy of this object.
- Returns
- copy
A copy of this object.
- get_fields(name)¶
Get all GenBank fields associated with a given field name.
- Parameters
- namestr
The field name.
- Returns
- fieldslist of (list of str, OrderedDict of str -> str)
A list containing the fields. For most field names, the list will only contain one element, but fields like REFERENCE are an exception. Each field is represented by a tuple. Each tuple contains as first element the content lines and as second element the subfields as dictionary. If the field has no subfields, the dictionary is empty.
- get_indices(name)¶
Get the indices to all GenBank fields associated with a given field name.
- Parameters
- namestr
The field name.
- Returns
- fieldslist of int
A list of indices. For most field names, the list will only contain one element, but fields like REFERENCE are an exception.
- insert(index, name, content, subfields=None)¶
Insert a GenBank field at the given position.
- Parameters
- indexint
The new field is inserted before the current field at this index. If the index is after the last field, the new field is appended to the end of the file.
- namestr
The field name.
- contentlist of str
The content lines.
- subfield_dictdict of str -> str, optional
The subfields of the field. The dictionary maps subfield names to the content lines of the respective subfield.
- classmethod read(file)¶
Read a GenBank file.
- Parameters
- filefile-like object or str
The file to be read. Alternatively a file path can be supplied.
- Returns
- file_objectGenBankFile
The parsed file.
- static read_iter(file)¶
Create an iterator over each line of the given text file.
- Parameters
- filefile-like object or str
The file to be read. Alternatively a file path can be supplied.
- Yields
- linestr
The current line in the file.
- set_field(name, content, subfield_dict=None)¶
Set a GenBank field with the given content.
If the field already exists in the file, the field is overwritten, otherwise a new field is created at the end of the file.
- Parameters
- namestr
The field name.
- contentlist of str
The content lines.
- subfield_dictdict of str -> str, optional
The subfields of the field. The dictionary maps subfield names to the content lines of the respective subfield.
- Raises
- InvalidFileError
If the field occurs multiple times in the file. In this case it is ambiguous which field to overwrite.
- write(file)¶
Write the contents of this object into a file (or file-like object).
- Parameters
- filefile-like object or str
The file to be written to. Alternatively a file path can be supplied.
- static write_iter(file, lines)¶
Iterate over the given lines of text and write each line into the specified file.
In contrast to
write(), each line of text is not stored in an intermediateTextFile, but is directly written to the file. Hence, this static method may save a large amount of memory if a large file should be written, especially if the lines are provided as generator.- Parameters
- filefile-like object or str
The file to be written to. Alternatively a file path can be supplied.
- linesgenerator or array-like of str
The lines of text to be written. Must not include line break characters.
Gallery¶
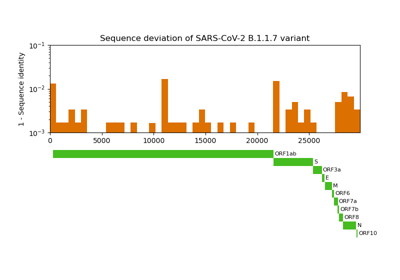
Comparative genome assembly of SARS-CoV-2 B.1.1.7 variant

Genome comparison between chloroplasts and cyanobacteria
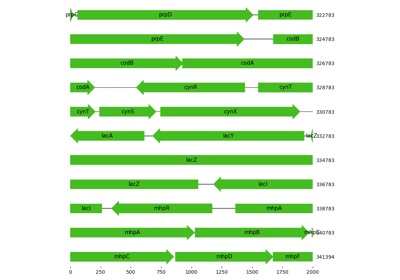
Visualization of region in proximity to lac operon
Three ways to get the secondary structure of a protein