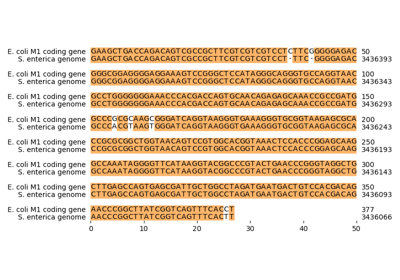
biotite.sequence.align.get_codes¶
- biotite.sequence.align.get_codes(alignment)[source]¶
Get the sequence codes of the sequences in the alignment.
The codes are built from the trace: Instead of the indices of the aligned symbols (trace), the return value contains the corresponding symbol codes for each index. Gaps are still represented by -1.
- Parameters
- alignmentAlignment
The alignment to get the sequence codes for.
- Returns
- codesndarray, dtype=int, shape=(n,m)
The sequence codes for the alignment. The shape is (n,m) for n sequences and m alignment cloumn. The array uses -1 values for gaps.
Examples
>>> seq1 = NucleotideSequence("CGTCAT") >>> seq2 = NucleotideSequence("TCATGC") >>> matrix = SubstitutionMatrix.std_nucleotide_matrix() >>> ali = align_optimal(seq1, seq2, matrix)[0] >>> print(ali) CGTCAT-- --TCATGC >>> print(get_codes(ali)) [[ 1 2 3 1 0 3 -1 -1] [-1 -1 3 1 0 3 2 1]]
Gallery¶
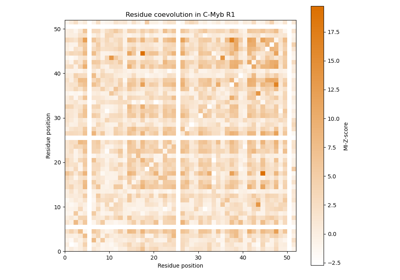
Mutual information as measure for coevolution of residues
Mutual information as measure for coevolution of residues