biotite.sequence.align.align_banded¶
- biotite.sequence.align.align_banded(seq1, seq2, matrix, band, gap_penalty=- 10, local=False, max_number=1000)[source]¶
Perform a local or semi-global alignment within a defined diagonal band. 1
The function requires two diagonals that defines the lower and upper limit of the alignment band. A diagonal is an integer defined as \(D = j - i\), where i and j are sequence positions in the first and second sequence, respectively. This means that two symbols at position i and j can only be aligned to each other, if \(D_L \leq j - i \leq D_U\). With increasing width of the diagonal band, the probability to find the optimal alignment, but also the computation time increases.
- Parameters
- seq1, seq2Sequence
The sequences to be aligned.
- matrixSubstitutionMatrix
The substitution matrix used for scoring.
- bandtuple(int, int)
The diagonals that represent the lower and upper limit of the search space. A diagonal \(D\) is defined as \(D = j-i\), where \(i\) and \(j\) are positions in seq1 and seq2, respectively. An alignment of sequence positions where \(D\) is lower than the lower limit or greater than the upper limit is not explored by the algorithm.
- gap_penaltyint or tuple(int, int), optional
If an integer is provided, the value will be interpreted as linear gap penalty. If a tuple is provided, an affine gap penalty is used. The first integer in the tuple is the gap opening penalty, the second integer is the gap extension penalty. The values need to be negative. (Default: -10)
- localbool, optional
If set to true, a local alignment is performed. Otherwise (default) a semi-global alignment is performed.
- max_numberint, optional
The maximum number of alignments returned. When the number of branches exceeds this value in the traceback step, no further branches are created.
- Returns
- alignmentslist of Alignment
The generated alignments. Each alignment in the list has the same similarity score, which is the maximum score possible within the defined band.
See also
align_optimalGuarantees to find the optimal alignment at the cost of greater compuation time and memory requirements.
Notes
The diagonals give the maximum difference between the number of inserted gaps. This means for any position in the alignment, the algorithm will not consider inserting a gap into a sequence, if the first sequence has already
-band[0]more gaps than the second sequence or if the second sequence has alreadyband[1]more gaps than the first sequence, even if inserting additional gaps would yield a more optimal alignment. Considerations on how to find a suitable band width are discussed in 2.The restriction to a limited band is the central difference between the banded alignment heuristic and the optimal alignment algorithms 34. Those classical algorithms require \(O(m \cdot n)\) memory space and computation time for aligning two sequences with lengths \(m\) and \(n\), respectively. The banded alignment algorithm reduces both requirements to \(O(\min(m,n) \cdot (D_U - D_L))\).
Implementation details
The implementation is very similar to
align_optimal(). The most significant difference is that not the complete alignment table is filled, but only the cells that lie within the diagonal band. Furthermore, to reduce also the space requirements the diagnoal band is ‘straightened’, i.e. the table’s rows are indented to the left. Hence, this table.
.
x
x
x
.
.
.
.
.
.
.
.
x
x
x
.
.
.
.
.
.
.
.
x
x
x
.
.
.
.
.
.
.
.
x
x
x
.
.
.
.
.
.
.
.
x
x
x
.
is transformed into this table:
x
x
x
x
x
x
x
x
x
x
x
x
x
x
x
Filled cells, i.e. cells within the band, are indicated by
x. The shorter sequence is always represented by the first dimension of the table in this implementation.References
- 1
W. R. Pearson, D. J. Lipman, “Improved tools for biological sequence comparison.,” Proceedings of the National Academy of Sciences of the United States of America, vol. 85, pp. 2444–2448, April 1988. doi: 10.1073/pnas.85.8.2444
- 2
J. Gibrat, “A short note on dynamic programming in a band,” BMC Bioinformatics, vol. 19, pp. 226, June 2018. doi: 10.1186/s12859-018-2228-9
- 3
S. B. Needleman, C. D. Wunsch, “A general method applicable to the search for similarities in the amino acid sequence of two proteins,” Journal of Molecular Biology, vol. 48, pp. 443–453, March 1970. doi: 10.1016/0022-2836(70)90057-4
- 4
T. F. Smith, M. S. Waterman, “Identification of common molecular subsequences,” Journal of Molecular Biology, vol. 147, pp. 195–197, March 1981. doi: 10.1016/0022-2836(81)90087-5
Examples
Find a matching diagonal for two sequences:
>>> sequence1 = NucleotideSequence("GCGCGCTATATTATGCGCGC") >>> sequence2 = NucleotideSequence("TATAAT") >>> table = KmerTable.from_sequences(k=4, sequences=[sequence1]) >>> match = table.match(sequence2)[0] >>> diagonal = match[0] - match[2] >>> print(diagonal) -6
Align the sequences centered on the diagonal with buffer in both directions:
>>> BUFFER = 5 >>> matrix = SubstitutionMatrix.std_nucleotide_matrix() >>> alignment = align_banded( ... sequence1, sequence2, matrix, ... band=(diagonal - BUFFER, diagonal + BUFFER), gap_penalty=(-6, -1) ... )[0] >>> print(alignment) TATATTAT TATA--AT
Gallery¶
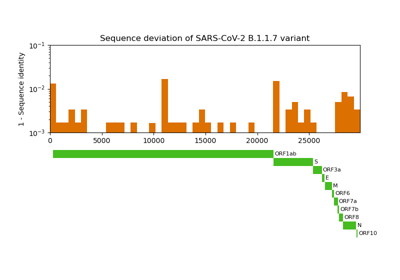
Comparative genome assembly of SARS-CoV-2 B.1.1.7 variant