biotite.sequence.Location¶
- class biotite.sequence.Location(first, last, strand=Strand.FORWARD, defect=Defect.NONE)[source]¶
Bases:
objectA
Locationdefines at which base(s)/residue(s) a feature is located.A feature can have multiple
Locationinstances if multiple locations are joined.Objects of this class are immutable.
- Attributes
- firstint
Starting base or residue position of the feature.
- lastint
Inclusive ending base or residue position of the feature.
- strandStrand
The strand direction. Always
Strand.FORWARDfor peptide features.- defectDefect
A possible defect of the location.
- class Defect(value)¶
Bases:
FlagThis enum type describes location defects.
A location has a defect, when the feature itself is not directly located in the range of the first to the last base.
NONE - No location defect
MISS_LEFT - A part of the feature has been truncated before the first base/residue of the
Location(probably by indexing anAnnotationobject)MISS_RIGHT - A part of the feature has been truncated after the last base/residue of the
Location(probably by indexing anAnnotationobject)BEYOND_LEFT - The feature starts at an unknown position before the first base/residue of the
LocationBEYOND_RIGHT - The feature ends at an unknown position after the last base/residue of the
LocationUNK_LOC - The exact position is unknown, but it is at a single base/residue between the first and last residue of the
Location, inclusiveBETWEEN - The position is between to consecutive bases/residues.
- NONE = 0¶
- MISS_LEFT = 1¶
- MISS_RIGHT = 2¶
- BEYOND_LEFT = 4¶
- BEYOND_RIGHT = 8¶
- UNK_LOC = 16¶
- BETWEEN = 32¶
Gallery¶
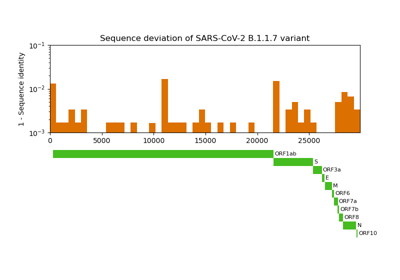
Comparative genome assembly of SARS-CoV-2 B.1.1.7 variant

Genome comparison between chloroplasts and cyanobacteria
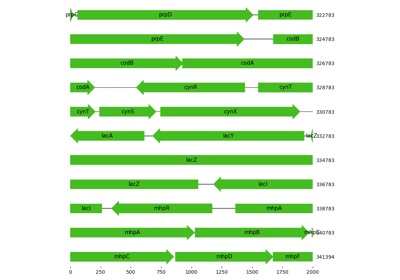
Visualization of region in proximity to lac operon
Three ways to get the secondary structure of a protein