Note
Go to the end to download the full example code
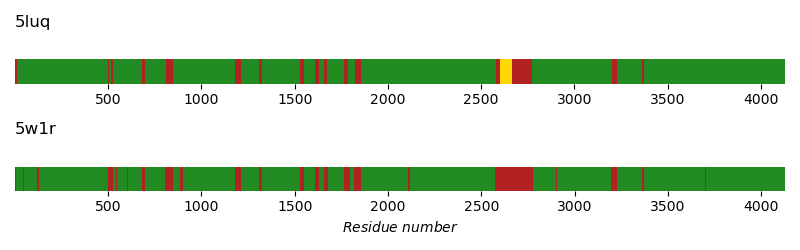
Missing residues in protein structures of DNA-PKcs¶
This script creates two bars, each indicating gaps in the structure of DNA-PKcs. The top bar indicates the missing residues in a recent crystal structure (PDB: 5LUQ), the bottom bar indicates missing residues in a recent cryo-EM structure (PDB: 5W1R).
Green: Space-resolved residues
Yellow: Residues with merely polyalanine annotation
Red: Unresolved residues
# Code source: Patrick Kunzmann
# License: BSD 3 clause
from tempfile import gettempdir
import biotite.structure.io as strucio
import biotite.database.rcsb as rcsb
import matplotlib.pyplot as plt
from matplotlib.patches import Rectangle
import numpy as np
def plot_gaps(pdb_id, chain_id, ax):
# Download and parse structure file
path = rcsb.fetch(pdb_id, "bcif", gettempdir())
atom_array = strucio.load_structure(path)
# Consider only one chain
atom_array = atom_array[atom_array.chain_id == chain_id]
# Array for saving the 'green', 'yellow' and 'red' state
states = np.zeros(atom_array.res_id[-1], dtype=int)
for i in range(len(states)):
# Get array for only one residue ID
residue = atom_array[atom_array.res_id == i+1]
if len(residue) == 0:
# not existing
states[i] = 0
elif residue.res_name[0] == "UNK":
# exisiting but polyalanine
states[i] = 1
else:
# existing
states[i] = 2
# Find the intervals for each state
state_intervals = []
curr_state = None
curr_start = None
for i in range(len(states)):
if curr_start is None:
curr_start = i
curr_state = states[i]
else:
if states[i] != states[i-1]:
state_intervals.append((curr_start, i, curr_state))
curr_start = i
curr_state = states[i]
state_intervals.append((curr_start, i, curr_state))
# Draw the state intervals as colored rectangles
for interval in state_intervals:
start = interval[0]
stop = interval[1]
state = interval[2]
if state == 0:
color = "firebrick"
elif state == 1:
color = "gold"
elif state == 2:
color = "forestgreen"
ax.add_patch(Rectangle((start+1-0.5, 0), stop-start, 1,
edgecolor="None", facecolor=color))
# Some other visual stuff
ax.spines["left"].set_visible(False)
ax.spines["bottom"].set_visible(False)
ax.spines["right"].set_visible(False)
ax.spines["top"].set_visible(False)
ax.yaxis.set_visible(False)
ax.set_xlim(0.5, len(states) + 0.5)
ax.set_ylim(0, 2)
fig = plt.figure(figsize=(8.0, 2.5))
ax = fig.add_subplot(211)
ax.set_title("5luq", loc="left")
plot_gaps("5luq", "A", ax)
ax = fig.add_subplot(212)
ax.set_title("5w1r", loc="left")
plot_gaps("5w1r", "A", ax)
ax.set_xlabel("$Residue \ number$")
fig.tight_layout()
plt.show()

