biotite.structure.AtomArray¶
- class biotite.structure.AtomArray(length)[source]¶
Bases:
_AtomArrayBaseAn array representation of a model consisting of multiple atoms.
An
AtomArraycan be seen as a list ofAtominstances. Instead of using directly a list, this class uses an NumPyndarrayfor each annotation category and the coordinates. These coordinates can be accessed directly via thecoordattribute. The annotations are accessed either via the category as attribute name or theget_annotation(),set_annotation()method. Usage of custom annotations is achieved viaadd_annotation()orset_annotation(). A detailed description of each annotation category can be viewed here.In order to get an an subarray of an
AtomArray, NumPy style indexing is used. This includes slices, boolean arrays, index arrays and even Ellipsis notation. Using a single integer as index returns a singleAtominstance.Inserting or appending an
AtomArrayto anotherAtomArrayis done with the ‘+’ operator. Only the annotation categories, which are existing in both arrays, are transferred to the new array.Optionally, an
AtomArraycan store chemical bond information via aBondListobject. It can be accessed using thebondsattribute. If no bond information is available,bondsisNone. Consequently the bond information can be removed from theAtomArray, by settingbondstoNone. When indexing theAtomArraythe atom indices in the associatedBondListare updated as well, hence the indices in theBondListwill always point to the same atoms. If twoAtomArrayinstances are concatenated, the resultingAtomArraywill contain the mergedBondListif at least one of the operands contains bond information.The
boxattribute contains the box vectors of the unit cell or the MD simulation box, respectively. Hence, it is a 3 x 3 ndarray with the vectors in the last dimension. If no box is provided, the attribute isNone. Setting theboxattribute toNonemeans removing the box from the atom array.- Parameters
- lengthint
The fixed amount of atoms in the array.
Examples
Creating an atom array from atoms:
>>> atom1 = Atom([1,2,3], chain_id="A") >>> atom2 = Atom([2,3,4], chain_id="A") >>> atom3 = Atom([3,4,5], chain_id="B") >>> atom_array = array([atom1, atom2, atom3]) >>> print(atom_array.array_length()) 3
Accessing an annotation array:
>>> print(atom_array.chain_id) ['A' 'A' 'B']
Accessing the coordinates:
>>> print(atom_array.coord) [[1. 2. 3.] [2. 3. 4.] [3. 4. 5.]]
NumPy style filtering:
>>> atom_array = atom_array[atom_array.chain_id == "A"] >>> print(atom_array.array_length()) 2
Inserting an atom array:
>>> insert = array([Atom([7,8,9], chain_id="C")]) >>> atom_array = atom_array[0:1] + insert + atom_array[1:2] >>> print(atom_array.chain_id) ['A' 'C' 'A']
- Attributes
- {annot}ndarray
Multiple n-length annotation arrays.
- coordndarray, dtype=float, shape=(n,3)
ndarray containing the x, y and z coordinate of the atoms.
- bondsBondList or None
A
BondList, specifying the indices of atoms that form a chemical bond.- boxndarray, dtype=float, shape=(3,3) or None
The surrounding box. May represent a MD simulation box or a crystallographic unit cell.
shapetuple of intTuple of array dimensions.
- add_annotation(category, dtype)¶
Add an annotation category, if not already existing.
Initially the new annotation is filled with the zero representation of the given type.
- Parameters
- categorystr
The annotation category to be added.
- dtypetype or str
A type instance or a valid NumPy dtype string. Defines the type of the annotation
See also
- array_length()¶
Get the length of the atom array.
This value is equivalent to the length of each annotation array. For
AtomArrayit is the same aslen(array).- Returns
- lengthint
Length of the array(s).
- copy()¶
Create a deep copy of this object.
- Returns
- copy
A copy of this object.
- del_annotation(category)¶
Removes an annotation category.
- Parameters
- categorystr
The annotation category to be removed.
- equal_annotation_categories(item)¶
Check, if this object shares equal annotation array categories with the given
AtomArrayorAtomArrayStack.- Parameters
- itemAtomArray or AtomArrayStack
The object to compare the annotation arrays with.
- Returns
- equalitybool
True, if the annotation array names are equal.
- equal_annotations(item)¶
Check, if this object shares equal annotation arrays with the given
AtomArrayorAtomArrayStack.- Parameters
- itemAtomArray or AtomArrayStack
The object to compare the annotation arrays with.
- Returns
- equalitybool
True, if the annotation arrays are equal.
- get_annotation(category)¶
Return an annotation array.
- Parameters
- categorystr
The annotation category to be returned.
- Returns
- arrayndarray
The annotation array.
- get_annotation_categories()¶
Return a list containing all annotation array categories.
- Returns
- categorieslist
The list containing the names of each annotation array.
- get_atom(index)¶
Obtain the atom instance of the array at the specified index.
The same as
array[index], if index is an integer.- Parameters
- indexint
Index of the atom.
- Returns
- atomAtom
Atom at position index.
- set_annotation(category, array)¶
Set an annotation array. If the annotation category does not exist yet, the category is created.
- Parameters
- categorystr
The annotation category to be set.
- arrayndarray or None
The new value of the annotation category. The size of the array must be the same as the array length.
Gallery¶
Mutual information as measure for coevolution of residues
Visualization of normal modes from an elastic network model
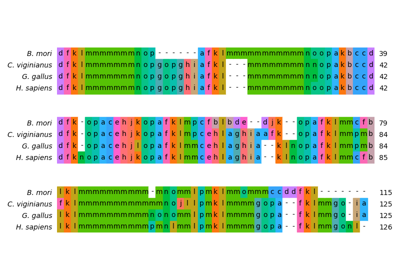
Structural alignment of lysozyme variants using ‘Protein Blocks’
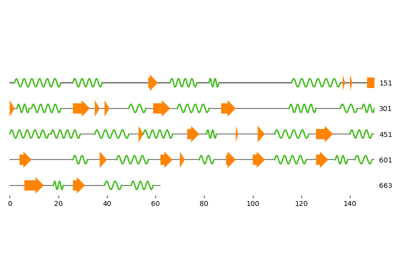
Three ways to get the secondary structure of a protein