biotite.sequence.Sequence¶
- class biotite.sequence.Sequence(sequence=())[source]¶
Bases:
CopyableThe abstract base class for all sequence types.
A
Sequencecan be seen as a succession of symbols, that are elements in the allowed set of symbols, theAlphabet. Internally, aSequenceobject uses a NumPyndarrayof integers, where each integer represents a symbol. TheAlphabetof aSequenceobject is used to encode each symbol, that is used to create theSequence, into an integer. These integer values are called symbol code, the encoding of an entire sequence of symbols is called sequence code.The size of the symbol code type in the array is determined by the size of the
Alphabet: If theAlphabetcontains 256 symbols or less, one byte is used per array element; if theAlphabetcontains between 257 and 65536 symbols, two bytes are used, and so on.Two
Sequenceobjects are equal if they are instances of the same class, have the sameAlphabetand have equal sequence codes. Comparison with a string or list of symbols evaluates always to false.A
Sequencecan be indexed by any 1-D index andarrayaccepts. If the index is a single integer, the decoded symbol at that position is returned, otherwise a subsequence is returned.Individual symbols of the sequence can also be exchanged in indexed form: If the an integer is used as index, the item is treated as a symbol. Any other index (slice, index list, boolean mask) expects multiple symbols, either as list of symbols, as
ndarraycontaining a sequence code or anotherSequenceinstance. Concatenation of two sequences is achieved with the ‘+’ operator.Each subclass of
Sequenceneeds to overwrite the abstract methodget_alphabet(), which specifies the alphabet theSequenceuses.- Parameters
- sequenceiterable object, optional
The symbol sequence, the
Sequenceis initialized with. For alphabets containing single letter strings, this parameter may also be a :class`str` object. By default the sequence is empty.
Examples
Creating a DNA sequence from string and print the symbols and the code:
>>> dna_seq = NucleotideSequence("ACGTA") >>> print(dna_seq) ACGTA >>> print(dna_seq.code) [0 1 2 3 0] >>> print(dna_seq.symbols) ['A' 'C' 'G' 'T' 'A'] >>> print(list(dna_seq)) ['A', 'C', 'G', 'T', 'A']
Sequence indexing:
>>> print(dna_seq[1:3]) CG >>> print(dna_seq[[0,2,4]]) AGA >>> print(dna_seq[np.array([False,False,True,True,True])]) GTA
Sequence manipulation:
>>> dna_copy = dna_seq.copy() >>> dna_copy[2] = "C" >>> print(dna_copy) ACCTA >>> dna_copy = dna_seq.copy() >>> dna_copy[0:2] = dna_copy[3:5] >>> print(dna_copy) TAGTA >>> dna_copy = dna_seq.copy() >>> dna_copy[np.array([True,False,False,False,True])] = "T" >>> print(dna_copy) TCGTT >>> dna_copy = dna_seq.copy() >>> dna_copy[1:4] = np.array([0,1,2]) >>> print(dna_copy) AACGA
Reverse sequence:
>>> dna_seq_rev = dna_seq.reverse() >>> print(dna_seq_rev) ATGCA
Concatenate the two sequences:
>>> dna_seq_concat = dna_seq + dna_seq_rev >>> print(dna_seq_concat) ACGTAATGCA
- Attributes
- codendarray
The sequence code.
- symbolslist
The list of symbols, represented by the sequence. The list is generated by decoding the sequence code, when this attribute is accessed. When this attribute is modified, the new list of symbols is encoded into the sequence code.
- alphabetAlphabet
The alphabet of this sequence. Cannot be set. Equal to get_alphabet().
- copy(new_seq_code=None)¶
Copy the object.
- Parameters
- new_seq_codendarray, optional
If this parameter is set, the sequence code is set to this value, rather than the original sequence code.
- Returns
- copy
A copy of this object.
- static dtype(alphabet_size)¶
Get the sequence code dtype required for the given size of the alphabet.
- abstract get_alphabet()¶
Get the
Alphabetof theSequence.This method must be overwritten, when subclassing
Sequence.- Returns
- alphabetAlphabet
Sequencealphabet.
- get_symbol_frequency()¶
Get the number of occurences of each symbol in the sequence.
If a symbol does not occur in the sequence, but it is in the alphabet, its number of occurences is 0.
- Returns
- frequencydict
A dictionary containing the symbols as keys and the corresponding number of occurences in the sequence as values.
- is_valid()¶
Check, if the sequence contains a valid sequence code.
A sequence code is valid, if at each sequence position the code is smaller than the size of the alphabet.
Invalid code means that the code cannot be decoded into symbols. Furthermore invalid code can lead to serious errors in alignments, since the substitution matrix is indexed with an invalid index.
- Returns
- validbool
True, if the sequence is valid, false otherwise.
- reverse(copy=True)¶
Reverse the
Sequence.- Parameters
- copybool, optional
If set to False, the code
ndarrayof the returned sequence is an array view to the sequence code of this object. In this case, manipulations on the returned sequence would also affect this object. Otherwise, the sequence code is copied.
- Returns
- reversedSequence
The reversed
Sequence.
Examples
>>> dna_seq = NucleotideSequence("ACGTA") >>> dna_seq_rev = dna_seq.reverse() >>> print(dna_seq_rev) ATGCA
Gallery¶
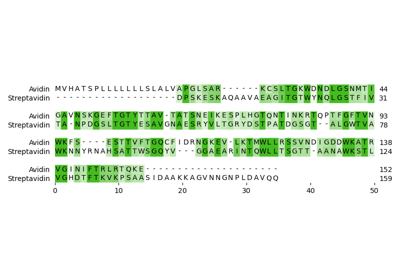
Pairwise sequence alignment of Avidin with Streptavidin
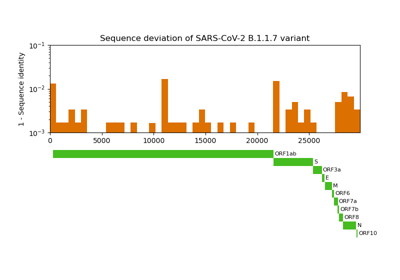
Comparative genome assembly of SARS-CoV-2 B.1.1.7 variant

Plot epitope mapping data onto protein sequence alignments
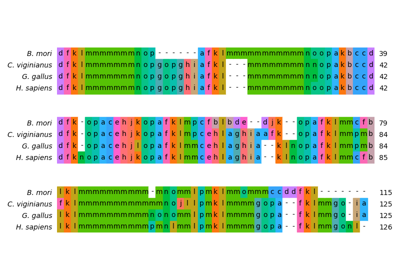
Structural alignment of lysozyme variants using ‘Protein Blocks’