biotite.sequence.io.genbank.get_locus¶
- biotite.sequence.io.genbank.get_locus(gb_file)[source]¶
Parse the LOCUS field of a GenBank or GenPept file.
- Parameters
- gb_fileGenBankFile
The GenBank file to read the LOCUS field from.
- Returns
- namestr
The locus name.
- lengthint
Sequence length.
- mol_typestr, optional
The molecule type. Usually one of
'DNA','RNA','Protein'or''.- is_circularbool, optional
True, if the sequence is circular, false otherwise.
- divisionstr, optional
The GenBank division to which the file belongs.
- datestr, optional
The date of last modification.
Examples
>>> import os.path >>> file = GenBankFile.read(os.path.join(path_to_sequences, "ec_bl21.gb")) >>> name, length, mol_type, is_circular, division, date = get_locus(file) >>> print(name) CP001509 >>> print(length) 4558953 >>> print(mol_type) DNA >>> print(is_circular) True >>> print(division) BCT >>> print(date) 16-FEB-2017
Gallery¶
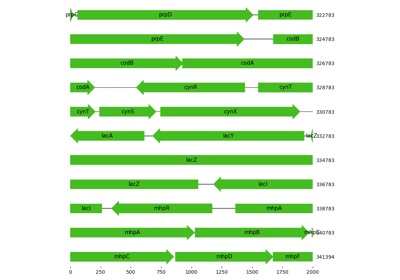
Visualization of region in proximity to lac operon
Visualization of region in proximity to lac operon
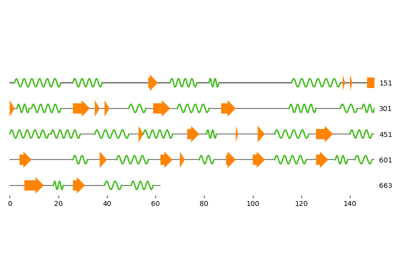
Three ways to get the secondary structure of a protein
Three ways to get the secondary structure of a protein