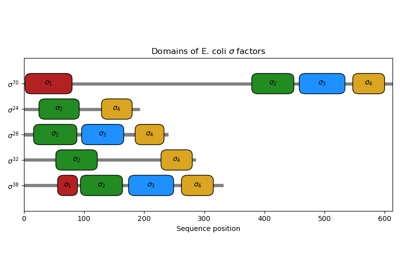
biotite.sequence.Feature¶
- class biotite.sequence.Feature(key, locs, qual=None)[source]¶
Bases:
CopyableThis class represents a single sequence feature, for example from a GenBank feature table. A feature describes a functional part of a sequence. It consists of a feature key, describing the general class of the feature, at least one location, describing its position on the reference, and qualifiers, describing the feature in detail.
Objects of this class are immutable.
- Parameters
- keystr
The name of the feature class, e.g. gene, CDS or regulatory.
- locsiterable object of Location
A list of feature locations. In most cases this list will only contain one location, but multiple ones are also possible for example in eukaryotic CDS (due to splicing).
- qualdict, optional
Maps feature qualifiers to their corresponding values. The keys are always strings. A value is either a string or
Noneif the qualifier key do not has a value. If key has multiple values, the values are separated by a line break.
- Attributes
- keystr
The name of the feature class, e.g. gene, CDS or regulatory.
- locsiterable object of Location
A list of feature locations. In most cases this list will only contain one location, but multiple ones are also possible for example in eukaryotic CDS (due to splicing).
- qualdict
Maps feature qualifiers to their corresponding values. The keys are always strings. A value is either a string or
Noneif the qualifier key do not has a value. If key has multiple values, the values are separated by a line break.
- copy()¶
Create a deep copy of this object.
- Returns
- copy
A copy of this object.
- get_location_range()¶
Get the minimum first base/residue and maximum last base/residue of all feature locations.
This can be used to create a location, that spans all of the feature’s locations.
- Returns
- firstint
The minimum first base/residue of all locations.
- lastint
The maximum last base/residue of all locations.
Gallery¶
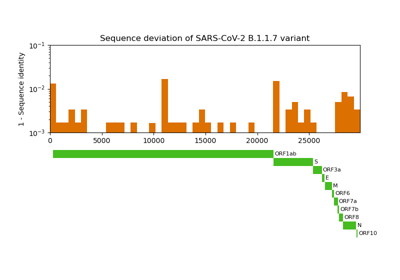
Comparative genome assembly of SARS-CoV-2 B.1.1.7 variant

Genome comparison between chloroplasts and cyanobacteria
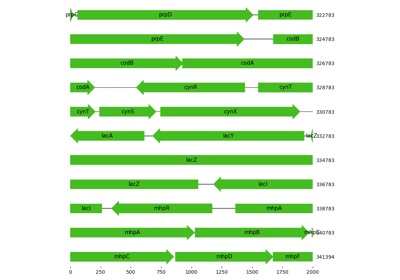
Visualization of region in proximity to lac operon
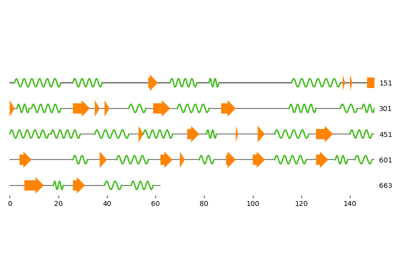
Three ways to get the secondary structure of a protein