biotite.application.blast.BlastAlignment¶
- class biotite.application.blast.BlastAlignment(sequences, trace, score, e_value, query_interval, hit_interval, hit_id, hit_definition)[source]¶
Bases:
AlignmentA specialized
Alignmentclass for alignments using the BLAST application. It stores additional data, like the E-value, the HSP position and a description of the hit sequence.Like its superclass, all attributes of a
BlastAlignmentare public. The attributes are the same as the constructor parameters.- Parameters
- sequenceslist
A list of aligned sequences. Does actually not contain the complete original sequences, but the HSP sequences.
- tracendarray, dtype=int, shape=(n,m)
The alignment trace.
- scoreint
Alignment score.
- e_valuefloat
Expectation value for the number of random sequences of a similar sized database getting an equal or higher score by change when aligned with the query sequence.
- query_intervaltuple of int
Describes the position of the HSP part of the query sequence in the original query sequence. The first element is the start position, the second element is the inclusive stop position. Indexing starts at 1.
- hit_intervaltuple of int
Analogous to query_interval, this describes the position of the HSP part of the hit sequence in the complete hit sequence.
- hit_idstr
The NCBI unique identifier (UID) of the hit sequence.
- hit_definitionstr
The name of the hit sequence.
- get_gapped_sequences()¶
Get a the string representation of the gapped sequences.
- Returns
- sequenceslist of str
The list of gapped sequence strings. The order is the same as in Alignment.sequences.
- static trace_from_strings(seq_str_list)¶
Create a trace from strings that represent aligned sequences.
- Parameters
- seq_str_listlist of str
The strings, where each each one represents a sequence (with gaps) in an alignment. A
-is interpreted as gap.
- Returns
- tracendarray, dtype=int, shape=(n,2)
The created trace.
Gallery¶
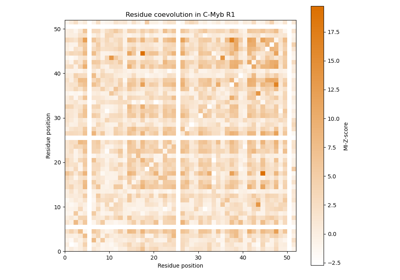
Mutual information as measure for coevolution of residues